The animation below represents a particular solution of the Navier-Stokes equations (1) and (2) and you will get it in the end of this problem¶
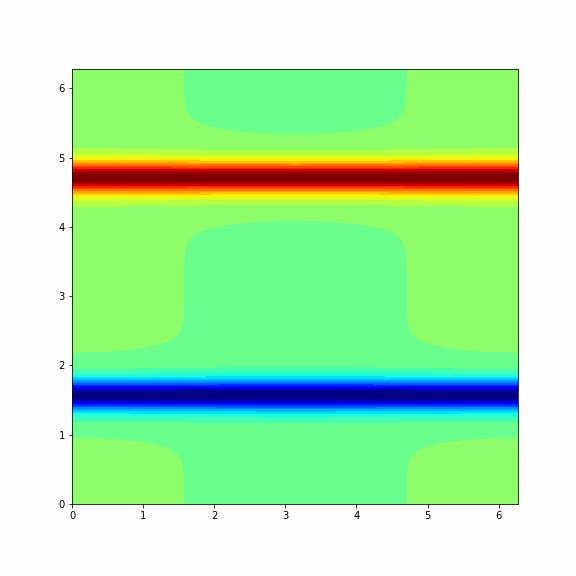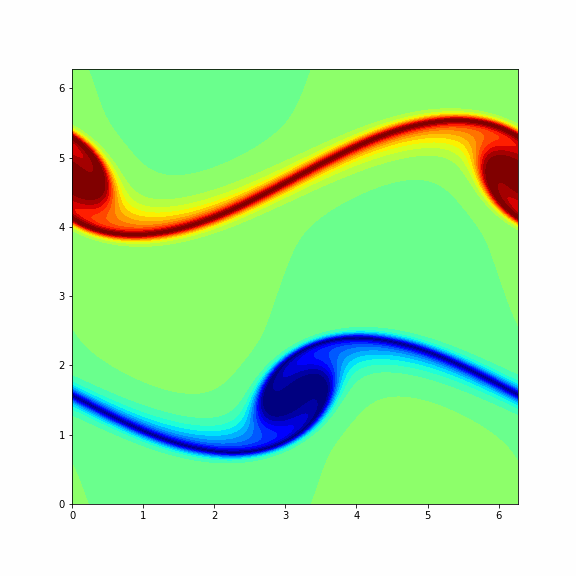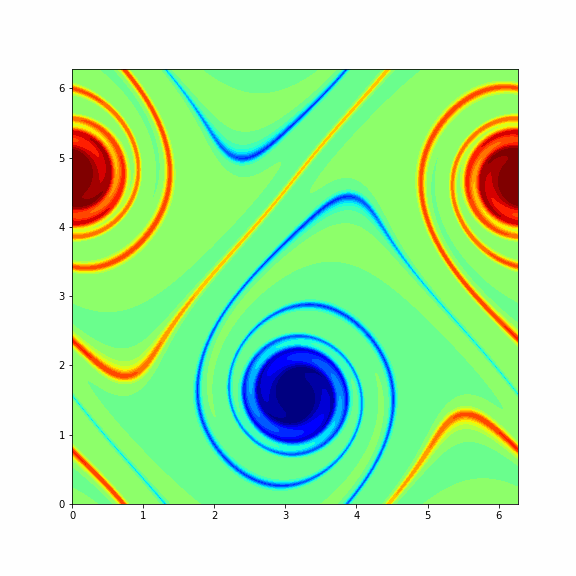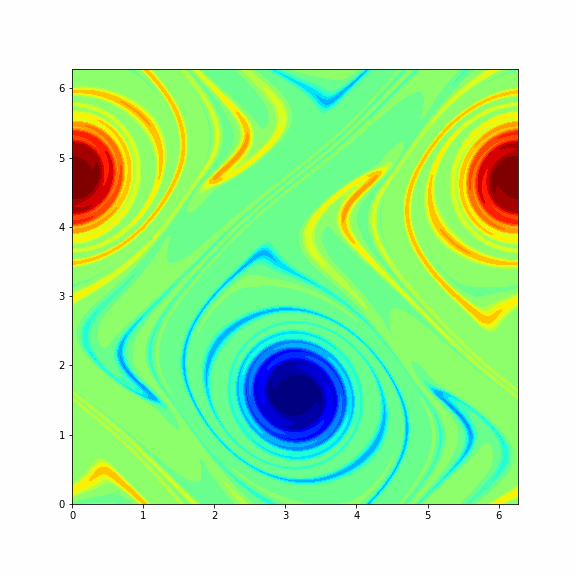
Note: To make a columnwise reshape in Python one should use np.reshape(X, order='f'), where the string 'f' stands for the Fortran ordering.
(2 pts) What is the complexity of a naive computation of $(A \otimes B) x$? Show how it can be reduced.
(3 pts) Let matrices $A$ and $B$ have eigendecompositions $A = S_A\Lambda_A S_A^{-1}$ and $B = S_B\Lambda_B S^{-1}_B$. Find eigenvectors and eigenvalues of the matrix $A\otimes I + I \otimes B$, where dimension of $I$ coincides with the dimension of $A$ and $B$.
(10 pts) Let $A = \mathrm{diag}\left(\frac{1}{1000},\frac{2}{1000},\dots \frac{999}{1000}, 1, 1000 \right)$. Estimate analytically the number of iterations required to solve linear system with $A$ with the relative accuracy $10^{-4}$ using
(5 pts) Provide numerical confirmation of your estimate from theoretical point of view
# Your solution is here
Given connected graph $G$ and its corresponding graph Laplacian matrix $L = D - A$ with eigenvalues $0=\lambda_1, \lambda_2, ..., \lambda_n$, where $D$ is its degree matrix and $A$ is its adjacency matrix, Fiedler vector is an eignevector correspondng to the second smallest eigenvalue $\lambda_2$ of $L$. Fiedler vector can be used for graph partitioning: positive values correspond to the one part of a graph and negative values to another.
To find the Fiedler vector we will use the inverse iteration with adaptive shifts (Rayleigh quotient iteration).
(5 pts) Write down the orthoprojection matrix on the space orthogonal to the eigenvector of $L$, corresponding to the eigenvalue $0$ and prove (analytically) that it is indeed an orthoprojection.
(5 pts) Implement the spectral partitioning as the function partition:
# INPUT:
# A - adjacency matrix (scipy.sparse.csr_matrix)
# num_iter_fix - number of iterations with fixed shift (int)
# shift - (float number)
# num_iter_adapt - number of iterations with adaptive shift (int) -- Rayleigh quotient iteration steps
# x0 - initial guess (1D numpy.ndarray)
# OUTPUT:
# x - normalized Fiedler vector (1D numpy.ndarray)
# eigs - eigenvalue estimations at each step (1D numpy.ndarray)
# eps - relative tolerance (float)
def partition(A, shift, num_iter_fix, num_iter_adapt, x0, eps):
x = x0
eigs = np.array([0])
return x, eigs
Algorithm must halt before num_iter_fix + num_iter_adapt iterations if the following condition is satisfied $$ \boxed{\|\lambda_k - \lambda_{k-1}\|_2 / \|\lambda_k\|_2 \leq \varepsilon} \text{ at some step } k.$$
Do not forget to use the orthogonal projection from above in the iterative process to get the correct eigenvector.
It is also a good idea to use shift=0 before the adaptive stragy is used. This, however, is not possible since the matrix $L$ is singular, and sparse decompositions in scipy does not work in this case. Therefore, we first use a very small shift instead.
(3 pts) Generate a random lollipop_graph using networkx library and find its partition. Draw this graph with vertices colored according to the partition.
(2 pts) Start the method with a random initial guess x0, set num_iter_fix=0 and comment why the method can converge to a wrong eigenvalue.
networkx, one of them should be $C_{30}$ - simple cyclic graph, and one of them should be $K_{30}$ - complete graph. (You also can change the number of vertices if it makes sense for your experiments, but do not make it trivially small).Let us deal here with a graph constructed from a binarized image. Consider the rule, that graph vertices are only pixels with $1$, and each vertex can have no more than $8$ connected vertices (pixel neighbours), $\textit{i.e}$ graph degree is limited by 8.
# Your solution is here
Disclaimer: this problem is released first time, so some typos can be found.
The governing equations for two-dimensional incompressible flows can be written in a dimensionless form as:
\begin{equation}\tag{1} \dfrac{\partial \omega}{\partial t} = \dfrac{1}{Re} \big(\dfrac{\partial^2 \omega}{\partial x^2} + \dfrac{\partial^2 \omega}{\partial y^2}\big) - \big(\dfrac{\partial \psi}{\partial y} \dfrac{\partial \omega}{\partial x} - \dfrac{\partial \psi}{\partial x} \dfrac{\partial \omega}{\partial y}\big), \end{equation}along with the kinematic relationship between vorticity $\omega(x,y,t)$ and stream function $\psi(x,y,t)$ according to the Poisson equation, which is given as:
\begin{equation}\tag{2} \dfrac{\partial^2 \psi}{\partial x^2} + \dfrac{\partial^2 \psi}{\partial y^2} = -\omega. \end{equation}We consider equations (1) and (2) in the computational domain $\Omega = [0, 2\pi] \times [0, 2\pi]$ and impose the following periodic boundary conditions:
$$\omega(x,0,t) =\omega(x, 2\pi, t), \quad \omega(0,y,t) =\omega(2\pi, y, t), \quad t \geq 0,$$and the same for $\psi(x,y,t)$.
Note: the Reynolds number, referred to as $Re$, is a fundamental physical constant that in particular determines whether the fluid flow is laminar or turbulent.

Fourier series expansion based methods are often used for solving problems with periodic boundary conditions. One of the most accurate methods for solving the Navier–Stokes equations in periodic domains is the pseudospectral method, which exploits the Fast Fourier Transform (FFT) algorithm.
Outline: the main idea of spectral methods is to write the solution of a differential equation as a sum of certain "basis functions" (e.g. Fourier series, Chebyshev polynomials etc) and then to choose the coefficients in the sum in order to satisfy the differential equation as well as possible.
Comprehensive survey of such methods can be found in this book.
We discretize the domain $[0,L_x]\times[0, L_y]$ by introducing a computation grid consisting of $N_x \times N_y$ equally spaced points.
The discrete grid coordinates for $i = 0, 1, \ldots, N_x$ and $j = 0, 1, \ldots, N_y$ are given by:
$$x_i = \frac{i L_x}{N_x}, \quad y_j = \frac{j L_y}{N_y}.$$Note, that since the domain is periodic $x_0 = x_{N_x}$ and $y_0 = y_{N_y}$.
Then, any discrete function $u_{i,j} = u(x_i,y_j)$ can be transformed to the Fourier space using the Discrete Fourier Transform (DFT):
$$ \tilde{u}_{m,n} = \sum_{i = 0}^{N_x - 1}\sum_{j = 0}^{N_y - 1} u_{i, j}e^{- \mathbf{i}(\frac{2\pi m}{L_x}x_i + \frac{2\pi n}{L_y}y_j)},$$and its inverse transform is:
$$ u_{i,j} = \frac{1}{N_x N_y} \sum_{m = -\frac{N_x}{2}}^{\frac{N_x}{2} - 1}\sum_{n = -\frac{N_y}{2}}^{\frac{N_y}{2} - 1} \tilde{u}_{m, n}e^{\mathbf{i}(\frac{2\pi m}{L_x}x_i + \frac{2\pi n}{L_y}y_j)},$$where $i$ and $j$ represent indices for the physical space (i.e. coordinates in the introduced grid), $m$ and $n$ are indices in the Fourier space (i.e. frequencies).
We also introduce wavenumbers:
$$k_x = \frac{2\pi m}{L_x}, \quad k_y = \frac{2 \pi n}{L_y}.$$Bonus question: how DFT coefficients $\tilde{u}_{m,n}$ relate to coefficients in the truncated Fourier series of $u(x,y)$?
In Fourier space we can easily perform differentiation with respect to $x$ and $y$. For example, the first and the second order derivatives of any function $u$ in discrete domain becomes:
$$ \left(\dfrac{\partial u}{\partial x}\right)_{i,j} = \frac{1}{N_x N_y}\sum_{m = -\frac{N_x}{2}}^{\frac{N_x}{2} - 1}\sum_{n = \frac{N_y}{2}}^{\frac{N_y}{2} - 1} \tilde{u}_{m, n} (\mathbf{i}k_x) e^{\mathbf{i}(k_x x_i + k_y y_j)}, $$$$ \left(\dfrac{\partial^2 u}{\partial x^2}\right)_{i,j} = \frac{1}{N_x N_y}\sum_{m = -\frac{N_x}{2}}^{\frac{N_x}{2} - 1}\sum_{n = -\frac{N_y}{2}}^{\frac{N_y}{2} - 1} \tilde{u}_{m, n} (-k_x^2) e^{\mathbf{i}(k_x x_i + k_y y_j)}, $$and similarly for the derivatives w.r.t. $y$
Assume $L_x = L_y = L = 2\pi$, $N_x = N_y = N$ for simplicity. Then, differentiation $\frac{\partial}{\partial x}$ in the Fourier space can be implemented as follows:
import numpy as np
import matplotlib.pyplot as plt
def dudx(u_tilde, N):
k1d = np.fft.fftfreq(N) * N
return u_tilde * (1j * k1d)
Note, we use np.fft.fftfreq(N) to determine the order of frequencies for certain numpy implementation (see the documentation of numpy.fft module for details).
Consider the following example:
L = 2*np.pi # size of computational domain
d = 7
N = 2**d
# discretize the domain $[0, 2\pi] \times [0, 2\pi]$ with uniform grid
ls = np.linspace(0, L, N, endpoint=False)
xx, yy = np.meshgrid(ls, ls, indexing='xy')
# define simple periodic function
u = np.sin(xx) * np.sin(yy)
# first, compute du/dx analytically
u_x = np.cos(xx) * np.sin(yy)
# next, compute du/dx in Fourier space
u_tilde = np.fft.fft2(u)
u_tilde_x = dudx(u_tilde, N)
u_x_fourier = np.fft.ifft2(u_tilde_x)
# check the result
err = np.linalg.norm(u_x - u_x_fourier)
print("error = ", err)
error = 5.437108258986234e-13
dudx(u_tilde, N) given above, your first task is to implement other derivatives arising in the Navier-Stokes equtions (1), (2). Loops are prohibited!def dudy(u_tilde, N):
pass
def d2udx2(u_tilde, N):
pass
def d2udy2(u_tilde, N):
pass
After transforming Eq. (1) and Eq. (2) to the Fourier space, the governing equations become:
\begin{equation}\tag{3} \frac{\partial \tilde{\omega}_{m,n}}{\partial t} = \frac{1}{Re}[(-k_x^2 - k_y^2)\tilde{\omega}_{m,n}] - \tilde{N}, \end{equation}\begin{equation}\tag{4} (-k_x^2 - k_y^2)\tilde{\psi}_{m,n} = -\tilde{\omega}_{m,n}, \end{equation}where $\tilde{N}$ represents the non-linear term which is computed using 2D convolutions as follows:
$$\tilde{N} = (\mathbf{i}k_y \tilde{\psi}_{m,n}) \circ (\mathbf{i}k_x \tilde{\omega}_{m,n}) - (\mathbf{i}k_x \tilde{\psi}_{m,n}) \circ (\mathbf{i}k_y \tilde{\omega}_{m,n}),$$i.e. multiplications in physical space become convolutions in the Fourier space.
To clarify where these convolutions come from, consider two discrete functions $u$ and $v$ represented by their DFT (1D for simplicity):
$$ u_{i} = \frac{1}{N_x} \sum_{m = -\frac{N_x}{2}}^{\frac{N_x}{2} - 1} \tilde{u}_{m}e^{\mathbf{i}\frac{2\pi m}{L_x}x_i},$$$$ v_{i} = \frac{1}{N_x} \sum_{n = -\frac{N_x}{2}}^{\frac{N_x}{2} - 1}\tilde{v}_{n}e^{\mathbf{i}\frac{2\pi n}{L_x}x_i}.$$Then, the direct multiplication results in: $$ u_{i} v_{i} = \frac{1}{N_x} \sum_{k = -N_x}^{N_x - 2} \frac{1}{N_x}\tilde{w}_{k}e^{\mathbf{i}\frac{2\pi k}{L_x}x_i},$$ where the coefficients $\tilde{\omega}_k$ are computed as follows (check it!):
$$\tilde{w}_{k} = \sum_{m + n = k}\tilde{u}_m\tilde{v}_n.$$Below we provide a possible implementation of 2D convolution using scipy.signal module. Note, that full convolution introduces higher frequinces that should be truncated in a proper way.
from scipy import signal
def conv2d_scipy(u_tilde, v_tilde, N):
# np.fft.fftshift is used to align implementation and formulas
full_conv = signal.convolve(np.fft.fftshift(u_tilde),\
np.fft.fftshift(v_tilde), mode='full')
trunc_conv = full_conv[N//2:-N//2+1, N//2:-N//2+1]
return np.fft.ifftshift(trunc_conv)/(N*N)
(10 pts) Your second task is to implement the same 2D convolution but using the Convolution Theorem in this time.
Hint: From the lecture course you should know that applying Convolution Theorem is straightforward when computing circular (or periodic) convolutions. However, for this task you should use an appropriate zero-padding by a factor of two (with further truncation).
def conv2d(u_tilde, v_tilde, N):
pass
# check yourself
u_tilde = np.random.rand(N, N)
v_tilde = np.random.rand(N, N)
err = np.linalg.norm(conv2d(u_tilde, v_tilde, N) - conv2d_scipy(u_tilde, v_tilde, N))
print("error =", err) # should be close to machine precision
Poisson solver
Finally, we need to solve the Poisson equation Eq. (2) which can be easily computed in the Fourier space according to the Eq. (4).
(5 pts) Implement inverse of the laplacian operator according to the template provided below. Note: the laplacian operator with periodic boundary conditions is singular (since the constant function is in nullspace). So, in order to avoid division by zero:
def laplace_inverse(omega_tilde, N):
psi_tilde = None
return psi_tilde
# check yourself
# consider simple solution
sol_analytic = np.sin(xx)*np.sin(yy)
# compute corresponding right hand side analytically
rhs = -2*np.sin(xx)*np.sin(yy)
# solve Poisson problem in Fourier space
rhs_tilde = np.fft.fft2(rhs)
sol_tilde = laplace_inverse(rhs_tilde, N)
sol = np.fft.ifft2(sol_tilde)
# check error is small
err = np.linalg.norm(sol - sol_analytic)
print("error =", err)
error = 1.8561658787461062e-14
Time integration
Eqs. (3) and (4) can be considered as semi-discrete ordinary differential equations (ODEs) obtained after (spectral) spatial discretization of the partial differential equations (1) and (2):
\begin{equation}\tag{5} \frac{d \tilde{\omega}}{dt} = \mathcal{L}(\tilde{\omega}, \tilde{\psi}), \end{equation}where $\mathcal{L}( \tilde{\omega} , \tilde{\psi})$ is the discrete operator of spatial derivatives including non-linear convective terms, linear diffusive terms, and $\tilde{\psi}$ which is obtained from the Poisson equation (4).
(5 pts) Implement $\mathcal{L}$ according to the template provided below
def L_op(omega_tilde, psi_tilde, N, Re=1):
pass
We integrate in time using fourth-order Runge–Kutta scheme that can be written in the following form:
$$\tilde{\omega}^{(1)} = \tilde{\omega}^{n} + \frac{\Delta t}{2}\mathcal{L}(\tilde{\omega}^{n}, \tilde{\psi}^{n})$$$$\tilde{\omega}^{(2)} = \tilde{\omega}^{n} + \frac{\Delta t}{2}\mathcal{L}(\tilde{\omega}^{(1)}, \tilde{\psi}^{(1)})$$$$\tilde{\omega}^{(3)} = \tilde{\omega}^{n} + \Delta t\mathcal{L}(\tilde{\omega}^{(2)}, \tilde{\psi}^{(2)})$$$$\tilde{\omega}^{n+1} = \frac{1}{3}(-\tilde{\omega}^{n} + \tilde{\omega}^{(1)} + 2\tilde{\omega}^{(2)} + \tilde{\omega}^{(3)}) + \frac{\Delta t}{6}\mathcal{L}(\tilde{\omega}^{3}, \tilde{\psi}^{3})$$def integrate_runge_kutta(omega0_tilde, N, n_steps, tau, Re):
omega_prev = omega0_tilde
psi_prev = laplace_inverse(-omega_prev, N)
for step in range(n_steps):
if(step%100 == 0):
print(step)
omega_1 = omega_prev + (tau/2)*L_op(omega_prev, psi_prev, N, Re)
psi_1 = -laplace_inverse(omega_1, N)
omega_2 = omega_prev + (tau/2)*L_op(omega_1, psi_1, N, Re)
psi_2 = -laplace_inverse(omega_2, N)
omega_3 = omega_prev + tau*L_op(omega_2, psi_2, N, Re)
psi_3 = -laplace_inverse(omega_3, N)
omega_next = (1./3)*(-omega_prev + omega_1 + 2*omega_2 + omega_3) + (tau/6)*L_op(omega_3, psi_3, N, Re)
psi_next = -laplace_inverse(omega_next, N)
omega_prev = omega_next
psi_prev = psi_next
return omega_prev
We first consider the Taylor-Green vortex (known analytical solution of the Navier-Stokes equations) to validate our solver:
# Taylor-Green vortex -- analytical solution for validation purposes
def taylor_green_vortex(xx, yy, t, N, Re):
k = 3
omega = 2*k*np.cos(k*xx)*np.cos(k*yy)*np.exp(-2*k**2*t*(1/Re))
return omega
Re = 1000
tau = 1e-2 # timestep
n_steps = 100
T = tau * n_steps # finial time
omega0 = taylor_green_vortex(xx, yy, 0, N, Re) # initial vorticity
omega0_tilde = np.fft.fft2(omega0) # convert to the Fourier space
omegaT_tilde = integrate_runge_kutta(omega0_tilde, N, n_steps, tau, Re) # integrate in time in the Fourier space
omegaT = np.real(np.fft.ifft2(omegaT_tilde)) # return back to physical space
0
# check the error is small
omegaT_analytical = taylor_green_vortex(xx, yy, T, N, Re)
err = np.linalg.norm(omegaT_analytical - omegaT)
print("error =", err)
error = 2.3043898350926834e-12
Finaly, we consider another (more interesting) initial vorticity that gives the dynamic from the GIF in the beginning of this problem.
# intial condition that evolves like a vortex
def shear_layer0(xx, yy, N):
delta = 0.05
sigma = 15/np.pi
a = delta*np.cos(yy[:, :N//2]) - sigma*(np.cosh(sigma*(xx[:, :N//2] - np.pi/2)))**(-2)
b = delta*np.cos(yy[:, N//2:]) + sigma*(np.cosh(sigma*(3*np.pi/2 - xx[:, N//2:])))**(-2)
return np.concatenate((a, b), axis=1)
Re = 10000
tau = 1e-3 # timestep
n_steps = 10000
T = tau * n_steps # finial time
omega0 = shear_layer0(xx, yy, N) # initial vorticity
omega0_tilde = np.fft.fft2(omega0) # convert to the Fourier space
omegaT_tilde = integrate_runge_kutta(omega0_tilde, N, n_steps, tau, Re) # integrate in time in the Fourier space
omegaT = np.real(np.fft.ifft2(omegaT_tilde)) # return back to physical space
0 100 200 300 400 500 600 700 800 900 1000 1100 1200 1300 1400 1500 1600 1700 1800 1900 2000 2100 2200 2300 2400 2500 2600 2700 2800 2900 3000 3100 3200 3300 3400 3500 3600 3700 3800 3900 4000 4100 4200 4300 4400 4500 4600 4700 4800 4900 5000 5100 5200 5300 5400 5500 5600 5700 5800 5900 6000 6100 6200 6300 6400 6500 6600 6700 6800 6900 7000 7100 7200 7300 7400 7500 7600 7700 7800 7900 8000 8100 8200 8300 8400 8500 8600 8700 8800 8900 9000 9100 9200 9300 9400 9500 9600 9700 9800 9900
# plot the solution at the final timestamp
plt.imshow(np.real(np.fft.ifft2(omega_final)), cmap='jet')
<matplotlib.image.AxesImage at 0x7f8c7646cb70>