Separator and block arrowhead structure: example¶
Separator for the 2D Laplacian matrix
$$ A_{2D} = I \otimes A_{1D} + A_{1D} \otimes I, \quad A_{1D} = \mathrm{tridiag}(-1, 2, -1), $$is as follows
Today we will sequentially discuss approaches to minimize fill-in
The idea is to eliminate rows and/or columns with fewer non-zeros, update fill-in and then repeat. How it relates to Markowitz pivoting?
Efficient implementation is an issue (adding/removing elements).
Current champion is "approximate minimal degree" by Amestoy, Davis, Duff.
It is suboptimal even for 2D PDE problems
SciPy sparse package uses minimal ordering approach for different matrices ($A^{\top}A$, $A + A^{\top}$)
Definition. A separator in a graph $G$ is a set $S$ of vertices whose removal leaves at least two connected components.
Separator $S$ gives the following ordering for an $N$-vertex graph $G$:
Separator for the 2D Laplacian matrix
$$ A_{2D} = I \otimes A_{1D} + A_{1D} \otimes I, \quad A_{1D} = \mathrm{tridiag}(-1, 2, -1), $$is as follows

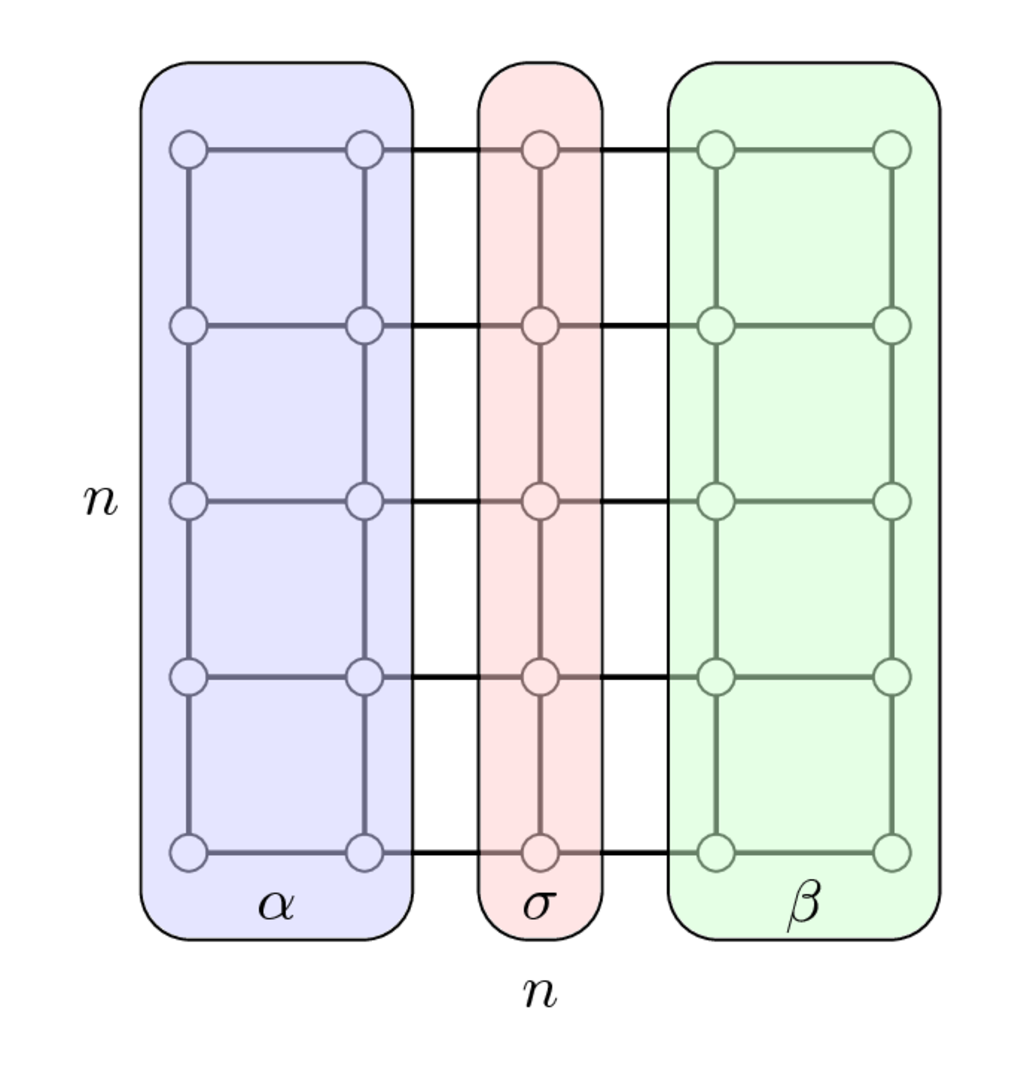
Once we have enumerated first indices in $\alpha$, then in $\beta$ and separators indices in $\sigma$ we get the following matrix
$$ PAP^\top = \begin{bmatrix} A_{\alpha\alpha} & & A_{\alpha\sigma} \\ & A_{\beta\beta} & A_{\beta\sigma} \\ A_{\sigma\alpha} & A_{\sigma\beta} & A_{\sigma\sigma}\end{bmatrix} $$which has arrowhrad structure.
For blocks $A_{\alpha\alpha}$, $A_{\beta\beta}$ we continue splitting recursively.
When the recursion is done, we need to eliminate blocks $A_{\sigma\alpha}$ and $A_{\sigma\beta}$.
This makes block in the position of $A_{\sigma\sigma}\in\mathbb{R}^{n\times n}$ dense.
Calculation of Cholesky of this block costs $\mathcal{O}(n^3) = \mathcal{O}(N^{3/2})$, where $N = n^2$ is the total number of nodes.
So, the complexity is $\mathcal{O}(N^{3/2})$
All of them have interfaces for C/C++, Fortran and Matlab
Computing separators is not a trivial task.
Graph partitioning heuristics has been an active research area for many years, often motivated by partitioning for parallel computation.
Existing approaches:
The idea of spectral partitioning goes back to Miroslav Fiedler, who studied connectivity of graphs (paper).
We need to split the vertices into two sets.
Consider +1/-1 labeling of vertices and the cost
and since we have +1/-1 labels, we have
$$\sum_i x^2_i = n \quad \Longleftrightarrow \quad \|x\|_2^2 = n.$$Cost $E_c$ can be written as (check why)
$$E_c = (Lx, x)$$where $L$ is the graph Laplacian, which is defined as a symmetric matrix with
$$L_{ii} = \mbox{degree of node $i$},$$$$L_{ij} = -1, \quad \mbox{if $i \ne j$ and there is an edge},$$and $0$ otherwise.
Minimization of $E_c$ with the mentioned constraints leads to a partitioning that tries to minimize number of edges in a separator, while keeping the partition balanced.
We now relax the integer quadratic programming to the continuous quadratic programming
Since $e$ is the eigenvector, corresponding to the smallest eigenvalue, on the space $x^\top e =0$ we get the second minimal eigevalue.
The sign $x_i$ indicates the partitioning.
In computations, we need to find out, how to find this second minimal eigenvalue –– we at least know about power method, but it finds the largest. We will discuss iterative methods for eigenvalue problems later in our course.
This is the main goal of the iterative methods for large-scale linear problems, and can be achieved via few matrix-by-vector products.
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
import networkx as nx
kn = nx.read_gml('karate.gml')
print("Number of vertices = {}".format(kn.number_of_nodes()))
print("Number of edges = {}".format(kn.number_of_edges()))
nx.draw_networkx(kn, node_color="red") #Draw the graph
Number of vertices = 34 Number of edges = 78
import scipy.sparse.linalg as spsplin
Laplacian = nx.laplacian_matrix(kn).asfptype()
plt.spy(Laplacian, markersize=5)
plt.title("Graph laplacian")
plt.axis("off")
plt.show()
eigval, eigvec = spsplin.eigsh(Laplacian, k=2, which="SM")
print("The 2 smallest eigenvalues =", eigval)
The 2 smallest eigenvalues = [-2.42470080e-16 4.68525227e-01]
plt.scatter(np.arange(len(eigvec[:, 1])), np.sign(eigvec[:, 1]))
plt.show()
print("Sum of elements in Fiedler vector = {}".format(np.sum(eigvec[:, 1].real)))
Sum of elements in Fiedler vector = 1.790234627208065e-15
nx.draw_networkx(kn, node_color=np.sign(eigvec[:, 1]))
Definition. The algebraic connectivity of a graph is the second-smallest eigenvalue of the Laplacian matrix.
Claim. The algebraic connectivity of a graph is greater than 0 if and only if a graph is a connected graph.
Computing bisection recursively is expensive.
As an alternative, one typically computes multilevel bisection that consists of 3 phases.
Once the permutation has been computed, we need to implement the elimination, making use of efficient computational kernels.
If in the elemination we will be able to get the elements into blocks, we will be able to use BLAS-3 computations.
It is done by supernodal data structures:
If adjacent rows have the same sparsity structure, they can be stored in blocks:
Also, we can use such structure in efficient computations!
Concept of iterative methods for linear systems:
If we want to achieve $\mathcal{O}(N)$ complexity of solving sparse linear systems, then direct solvers are not appropriate.
If we want to solve partial eigenproblem, the full eigendecomposition is too costly.
For both problems we will use iterative, Krylov subspace solvers, which treat the matrix as a black-box linear operator.
We have now an absolutely different view on a matrix: matrix is now a linear operator, that acts on a vector,
and this action can be computed in $\mathcal{O}(N)$ operations.
This is the only information we know about the matrix: the matrix-by-vector product (matvec)
Can we solve linear systems using only matvecs?
Of course, we can multiply by the colums of the identity matrix, and recover the full matrix, but it is not what we need.
The simplest idea is the "simple iteration method" or Richardson iteration.
$$Ax = f,$$ $$\tau (Ax - f) = 0,$$ $$x - \tau (Ax - f) = x,$$ $$x_{k+1} = x_k - \tau (Ax_k - f),$$
where $\tau$ is the iteration parameter, which can be always chosen such that the method converges.
which leads to the Richardson iteration
$$ y_{k+1} = y_k - \tau(Ay_k -f) $$therefore if $\Vert I - \tau A \Vert < 1$ in any norm, the iteration converges.
For symmetric positive definite case it is always possible to select $\tau$ such that the method converges.
What about the non-symmetric case? Below demo will be presented...
where $\lambda_{\min}$ is the minimal eigenvalue, and $\lambda_{\max}$ is the maximal eigenvalue of the matrix $A$.
Even with the optimal parameter choice, the error at the next step satisfies
$$\|e_{k+1}\|_2 \leq q \|e_k\|_2 , \quad\rightarrow \quad \|e_k\|_2 \leq q^{k} \|e_0\|_2,$$where
$$ q = \frac{\lambda_{\max} - \lambda_{\min}}{\lambda_{\max} + \lambda_{\min}} = \frac{\mathrm{cond}(A) - 1}{\mathrm{cond}(A)+1}, $$$$\mathrm{cond}(A) = \frac{\lambda_{\max}}{\lambda_{\min}} \quad \text{for} \quad A=A^*>0$$is the condition number of $A$.
Let us do some demo...
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
plt.rc("text", usetex=True)
import scipy as sp
import scipy.sparse
import scipy.sparse.linalg as spla
import scipy
from scipy.sparse import csc_matrix
n = 10
ex = np.ones(n);
A = sp.sparse.spdiags(np.vstack((-ex, 2*ex, -ex)), [-1, 0, 1], n, n, 'csr');
rhs = np.ones(n)
ev1, vec = spla.eigsh(A, k=2, which='LA')
ev2, vec = spla.eigsh(A, k=2, which='SA')
lam_max = ev1[0]
lam_min = ev2[0]
tau_opt = 2.0/(lam_max + lam_min)
fig, ax = plt.subplots()
plt.close(fig)
niters = 100
x = np.zeros(n)
res_richardson = []
for i in range(niters):
rr = A.dot(x) - rhs
x = x - tau_opt * rr
res_richardson.append(np.linalg.norm(rr))
#Convergence of an ordinary Richardson (with optimal parameter)
plt.semilogy(res_richardson)
plt.xlabel("Number of iterations, $k$", fontsize=20)
plt.ylabel("Residual norm, $\|Ax_k - b\|_2$", fontsize=20)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
print("Maximum eigenvalue = {}, minimum eigenvalue = {}".format(lam_max, lam_min))
cond_number = lam_max.real / lam_min.real
print("Condition number = {}".format(cond_number))
print(np.array(res_richardson)[1:] / np.array(res_richardson)[:-1])
print("Theoretical factor: {}".format((cond_number - 1) / (cond_number + 1)))
Maximum eigenvalue = 3.682507065662361, minimum eigenvalue = 0.08101405277100546 Condition number = 45.45516413147909 [0.9186479 0.94484203 0.95170654 0.95466793 0.95595562 0.95651542 0.9567591 0.95686534 0.95691172 0.95693198 0.95694084 0.95694472 0.95694642 0.95694716 0.95694748 0.95694763 0.95694769 0.95694771 0.95694773 0.95694773 0.95694773 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774 0.95694774] Theoretical factor: 0.9569477357923108
This is another reason why condition number is so important:
Main questions for the iterative method is how to make the matrix better conditioned.
Possible cases of Richardson iteration behaviour:
Q: how can we identify our case before running iterative method?
# B = np.random.randn(2, 2)
B = np.array([[1, 2], [-1, 0]])
# B = np.array([[0, 1], [-1, 0]])
x_true = np.zeros(2)
f = B.dot(x_true)
eigvals = np.linalg.eigvals(B)
print("Spectrum of the matrix = {}".format(eigvals))
# Run Richardson iteration
x = np.array([0, -1])
tau = 1e-2
conv_x = [x]
r = B.dot(x) - f
conv_r = [np.linalg.norm(r)]
num_iter = 1000
for i in range(num_iter):
x = x - tau * r
conv_x.append(x)
r = B.dot(x) - f
conv_r.append(np.linalg.norm(r))
Spectrum of the matrix = [0.5+1.32287566j 0.5-1.32287566j]
plt.semilogy(conv_r)
plt.xlabel("Number of iteration, $k$", fontsize=20)
plt.ylabel("Residual norm", fontsize=20)
Text(0, 0.5, 'Residual norm')
plt.scatter([x[0] for x in conv_x], [x[1] for x in conv_x])
plt.xlabel("$x$", fontsize=20)
plt.ylabel("$y$", fontsize=20)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.title("$x_0 = (0, -1)$", fontsize=20)
Text(0.5, 1.0, '$x_0 = (0, -1)$')
But before preconditioners, we can use better iterative methods.
There is a whole zoo of iterative methods, but we need to know just few of them.
This method is called the steepest descent.
However, it still converges similarly to the Richardson iteration.
Another way to find $\tau_k$ is to consider
$$e_{k+1} = (I - \tau_k A) e_k = (I - \tau_k A) (I - \tau_{k-1} A) e_{k-1} = \ldots = p(A) e_0, $$where $p(A)$ is a matrix polynomial (simplest matrix function)
$$ p(A) = (I - \tau_k A) \ldots (I - \tau_0 A), $$and $p(0) = 1$.
The error is written as
$$e_{k+1} = p(A) e_0, $$and hence
$$\|e_{k+1}\| \leq \|p(A)\| \|e_0\|, $$where $p(0) = 1$ and $p(A)$ is a matrix polynomial.
To get better error reduction, we need to minimize
$$\Vert p(A) \Vert$$over all possible polynomials $p(x)$ of degree $k+1$ such that $p(0)=1$. We will use $\|\cdot\|_2$.
Important special case: $A = A^* > 0$.
Then $A = U \Lambda U^*$,
and
$$\Vert p(A) \Vert_2 = \Vert U p(\Lambda) U^* \Vert_2 = \Vert p(\Lambda) \Vert_2 = \max_i |p(\lambda_i)| \overset{!}{\leq} \max_{\lambda_\min \leq \lambda {\leq} \lambda_\max} |p(\lambda)|.$$The latter inequality is the only approximation. Here we make a crucial assumption that we do not want to benefit from distribution of spectra between $\lambda_\min$ and $\lambda_\max$.
Thus, we need to find a polynomial such that $p(0) = 1$, that has the least possible deviation from $0$ on $[\lambda_\min, \lambda_\max]$.
We can do the affine transformation of the interval $[\lambda_\min, \lambda_\max]$ to the interval $[-1, 1]$:
$$ \xi = \frac{{\lambda_\max + \lambda_\min - (\lambda_\min-\lambda_\max)x}}{2}, \quad x\in [-1, 1]. $$The problem is then reduced to the problem of finding the polynomial least deviating from zero on an interval $[-1, 1]$.
The exact solution to this problem is given by the famous Chebyshev polynomials of the form
$$T_n(x) = \cos (n \arccos x)$$This is a polynomial!
We can express $T_n$ from $T_{n-1}$ and $T_{n-2}$:
$|T_n(x)| \leq 1$ on $x \in [-1, 1]$.
It has $(n+1)$ alternation points, where the maximal absolute value is achieved (this is the sufficient and necessary condition for the optimality) (Chebyshev alternance theorem, no proof here).
The roots are just
We can plot them...
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
x1 = np.linspace(-1, 1, 128)
x2 = np.linspace(-1.1, 1.1, 128)
p = np.polynomial.Chebyshev((0, 0, 0, 0, 0, 0, 0, 0, 0, 1), (-1, 1)) #These are Chebyshev series, a proto of "chebfun system" in MATLAB
fig, (ax1, ax2) = plt.subplots(1, 2)
ax1.plot(x1, p(x1))
ax1.set_title('Interval $x\in[-1, 1]$')
ax2.plot(x2, p(x2))
ax2.set_title('Interval $x\in[-1.1, 1.1]$')
Text(0.5, 1.0, 'Interval $x\\in[-1.1, 1.1]$')
Note that $p(x) = (1-\tau_n x)\dots (1-\tau_0 x)$, hence roots of $p(x)$ are $1/\tau_i$ and that we additionally need to map back from $[-1,1]$ to $[\lambda_\min, \lambda_\max]$. This results into
$$\tau_i = \frac{2}{\lambda_\max + \lambda_\min - (\lambda_\max - \lambda_\min)x_i}, \quad x_i = \cos \frac{\pi(2i + 1)}{2n}\quad i=0,\dots,n-1$$The convergence (we only give the result without the proof) is now given by
$$ e_{k+1} \leq C q^k e_0, \quad q = \frac{\sqrt{\mathrm{cond}(A)}-1}{\sqrt{\mathrm{cond}(A)}+1}, $$which is better than in the Richardson iteration.
import numpy as np
import matplotlib.pyplot as plt
A = sp.sparse.spdiags(np.vstack((-ex, 2*ex, -ex)), [-1, 0, 1], n, n, 'csr');
rhs = np.ones(n)
ev1, vec = spla.eigsh(A, k=2, which='LA')
ev2, vec = spla.eigsh(A, k=2, which='SA')
lam_max = ev1[0]
lam_min = ev2[0]
niters = 64
roots = [np.cos((np.pi * (2 * i + 1)) / (2 * niters)) for i in range(niters)]
taus = [(lam_max + lam_min - (lam_min - lam_max) * r) / 2 for r in roots]
x = np.zeros(n)
r = A.dot(x) - rhs
res_cheb = [np.linalg.norm(r)]
print(taus)
for i in range(niters):
x = x - 1.0 / taus[i] * r
r = A.dot(x) - rhs
res_cheb.append(np.linalg.norm(r))
plt.semilogy(res_richardson, label="Richardson")
plt.semilogy(res_cheb, label="Chebyshev")
plt.legend(fontsize=20)
plt.xlabel("Number of iterations, $k$", fontsize=20)
plt.ylabel("Residual norm, $\|Ax_k - b\|_2$", fontsize=20)
plt.xticks(fontsize=20)
_ = plt.yticks(fontsize=20)
[3.6819647144817464, 3.677627864992451, 3.6689646138641447, 3.6559958316276053, 3.638752761215172, 3.6172769426937834, 3.5916201131913166, 3.5618440822573167, 3.5280205829583746, 3.490231099066891, 3.448566668759529, 3.403127665298271, 3.354023555222442, 3.3013726346342267, 3.2453017442129988, 3.1859459636450125, 3.1234482862046082, 3.05795927427089, 2.9896366966097663, 2.918645148295174, 2.8451556541851284, 2.769345256907859, 2.691396590350614, 2.611497439678626, 2.5298402889442047, 2.4466218573757885, 2.3620426254641016, 2.2763063519870834, 2.189619583137152, 2.1021911549333296, 2.0142316901169863, 1.9259530907432036, 1.8375680276901654, 1.7492894283163825, 1.6613299635000391, 1.5739015352962171, 1.4872147664462856, 1.4014784929692676, 1.3168992610575803, 1.2336808294891646, 1.1520236787547429, 1.072124528082755, 0.9941758615255103, 0.9183654642482413, 0.8448759701381947, 0.7738844218236025, 0.7055618441624791, 0.640072832228761, 0.5775751547883565, 0.5182193742203698, 0.4621484837991421, 0.409497563210927, 0.36039345313509785, 0.3149544496738399, 0.27329001936647757, 0.23550053547499394, 0.20167703617605226, 0.17190100524205199, 0.14624417573958537, 0.12476835721819679, 0.10752528680576345, 0.09455650456922404, 0.08589325344091736, 0.08155640395162234]
niters = 64
roots = [np.cos((np.pi * (2 * i + 1)) / (2 * niters)) for i in range(niters)]
taus = [(lam_max + lam_min - (lam_min - lam_max) * r) / 2 for r in roots]
x = np.zeros(n)
r = A.dot(x) - rhs
res_cheb_even = [np.linalg.norm(r)]
print(taus)
# Implementation may be non-optimal if number of iterations is not power of two
def leb_shuffle_2n(n):
if n == 1:
return np.array([0,], dtype=int)
else:
prev = leb_shuffle_2n(n // 2)
ans = np.zeros(n, dtype=int)
ans[::2] = prev
ans[1::2] = n - 1 - prev
return ans
good_perm_even = leb_shuffle_2n(niters)
print(good_perm_even, len(good_perm_even))
# good_perm_even = np.random.permutation([i for i in range(niters)])
for i in range(niters):
x = x - 1.0/taus[good_perm_even[i]] * r
r = A.dot(x) - rhs
res_cheb_even.append(np.linalg.norm(r))
plt.semilogy(res_richardson, label="Richardson")
plt.semilogy(res_cheb_even, label="Chebyshev")
plt.legend(fontsize=20)
plt.xlabel("Number of iterations, $k$", fontsize=20)
plt.ylabel("Residual norm, $\|Ax_k - b\|_2$", fontsize=20)
plt.xticks(fontsize=20)
_ = plt.yticks(fontsize=20)
[3.6819647144817464, 3.677627864992451, 3.6689646138641447, 3.6559958316276053, 3.638752761215172, 3.6172769426937834, 3.5916201131913166, 3.5618440822573167, 3.5280205829583746, 3.490231099066891, 3.448566668759529, 3.403127665298271, 3.354023555222442, 3.3013726346342267, 3.2453017442129988, 3.1859459636450125, 3.1234482862046082, 3.05795927427089, 2.9896366966097663, 2.918645148295174, 2.8451556541851284, 2.769345256907859, 2.691396590350614, 2.611497439678626, 2.5298402889442047, 2.4466218573757885, 2.3620426254641016, 2.2763063519870834, 2.189619583137152, 2.1021911549333296, 2.0142316901169863, 1.9259530907432036, 1.8375680276901654, 1.7492894283163825, 1.6613299635000391, 1.5739015352962171, 1.4872147664462856, 1.4014784929692676, 1.3168992610575803, 1.2336808294891646, 1.1520236787547429, 1.072124528082755, 0.9941758615255103, 0.9183654642482413, 0.8448759701381947, 0.7738844218236025, 0.7055618441624791, 0.640072832228761, 0.5775751547883565, 0.5182193742203698, 0.4621484837991421, 0.409497563210927, 0.36039345313509785, 0.3149544496738399, 0.27329001936647757, 0.23550053547499394, 0.20167703617605226, 0.17190100524205199, 0.14624417573958537, 0.12476835721819679, 0.10752528680576345, 0.09455650456922404, 0.08589325344091736, 0.08155640395162234] [ 0 63 31 32 15 48 16 47 7 56 24 39 8 55 23 40 3 60 28 35 12 51 19 44 4 59 27 36 11 52 20 43 1 62 30 33 14 49 17 46 6 57 25 38 9 54 22 41 2 61 29 34 13 50 18 45 5 58 26 37 10 53 21 42] 64
We have made an important assumption about the spectrum: it is contained within an interval over the real line (and we need to know the bounds)
If the spectrum is contained within two intervals, and we know the bounds, we can also put the optimization problem for the optimal polynomial.
For the case of two segments the best polynomial is given by Zolotarev polynomials (expressed in terms of elliptic functions). Original paper was published in 1877, see details here
For the case of more than two segments the best polynomial can be expressed in terms of hyperelliptic functions
The implementation of the Chebyshev acceleration requires the knowledge of the spectrum.
It only stores the previous vector $x_k$ and computes the new correction vector
It belongs to the class of two-term iterative methods, i.e. it approximates $x_{k+1}$ using 2 vectors: $x_k$ and $r_k$.
It appears that if we store more vectors, then we can go without the spectrum estimation (and better convergence in practice)!
The Chebyshev method produces the approximation of the form
$$x_{k+1} = x_0 + p(A) r_0,$$i.e. it lies in the Krylov subspace of the matrix which is defined as
$$ \mathcal{K}_k(A, r_0) = \mathrm{Span}(r_0, Ar_0, A^2 r_0, \ldots, A^{k-1}r_0 ) $$The most natural approach then is to find the vector in this linear subspace that minimizes certain norm of the error
The idea is to minimize given functional:
To make methods practical one has to
We will consider these methods in details on the next lecture.
from IPython.core.display import HTML
def css_styling():
styles = open("./styles/custom.css", "r").read()
return HTML(styles)
css_styling()