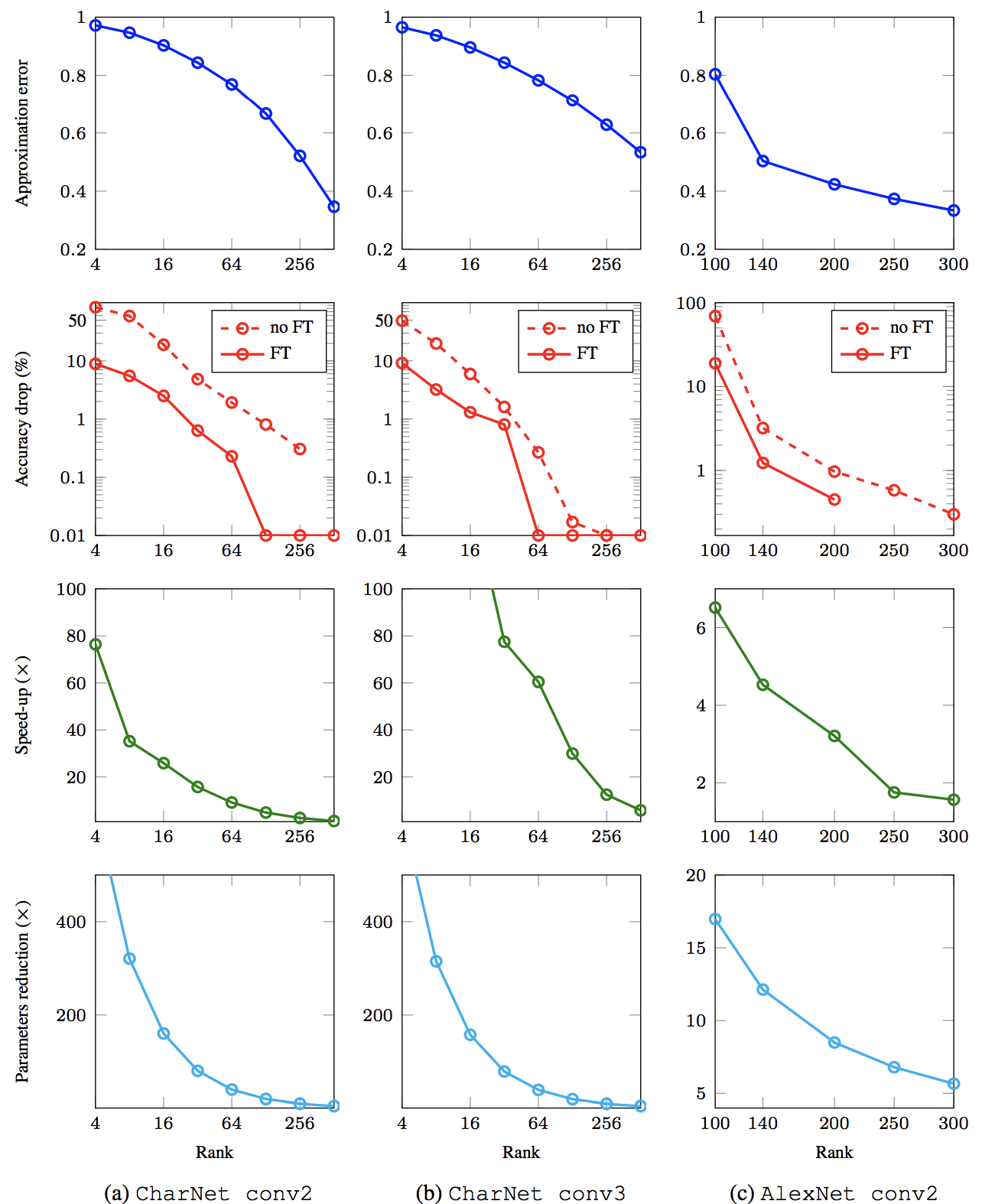
Tensors¶
By tensor we imply a multidimensional array:
$$ A(i_1, \dots, i_d), \quad 1\leq i_k\leq n_k, $$where $d$ is called dimension, $n_k$ - mode size. This is standard definition in applied mathematics community. For details see [1], [2], [3].
$d=2$ (matrices) $\Rightarrow$ classic theory (SVD, LU, QR, $\dots$)
$d\geq 3$ (tensors) $\Rightarrow$ under development. Generalization of standard matrix results is not straightforward.
Picture is taken from this presentation