Skeleton decomposition¶
A very useful representation for computation of the matrix rank is the skeleton decomposition and is closely related to the rank. This decompositions explains, why and how matrices of low rank can be compressed.
It can be graphically represented as follows:
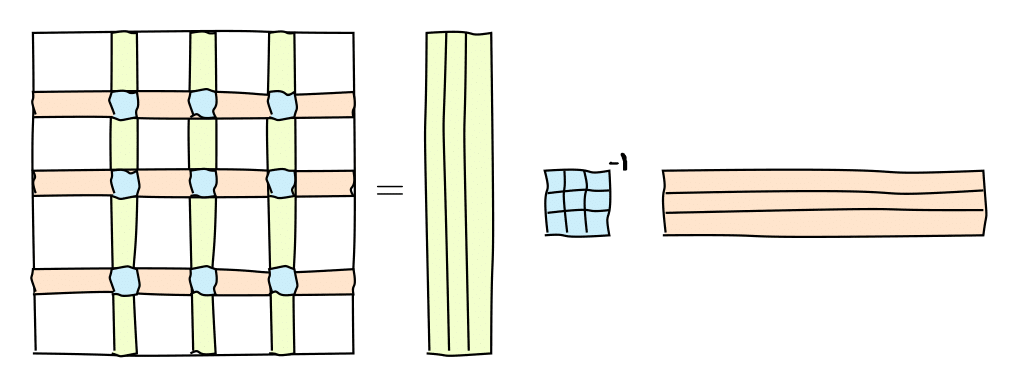 or in the matrix form
or in the matrix form
where $C$ are some $k=\mathrm{rank}(A)$ columns of $A$, $R$ are some $k$ rows of $A$ and $\widehat{A}$ is the nonsingular submatrix on the intersection.